In Get started you can find a description to get quick up-and-running on a Linux machine:
Setup a single Linux integration demo
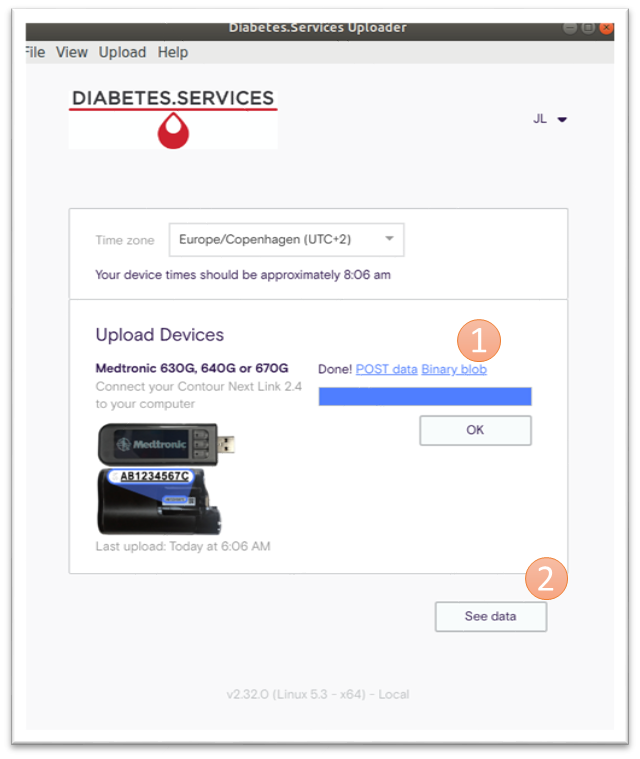
When finished uploading then you can do the following: (refer to the orange number in figure):
In the dataflow Data_Donation you can find a demonstration of these capabilities:
In the dataflow Data_Tracing you can find a demonstration of these capabilities:
We have defined a concept of a health data store (HDS) to merit of higher protection for sensitive data as health data. For scenarios of multiple users in the same environment we have developed a template
that can be used to demonstrate the workflow as swagger response.
In Proxy you can find a demonstration of these capabilities:

Dataflows